Volume 15, Issue 1 (2023)
Iran J War Public Health 2023, 15(1): 101-105 |
Back to browse issues page
Article Type:
Subject:
History
Received: 2022/11/26 | Accepted: 2023/03/8 | Published: 2023/03/16
Received: 2022/11/26 | Accepted: 2023/03/8 | Published: 2023/03/16
How to cite this article
Banoon S, Hussein Ali Z, Al-Kraety I, Aziz Z. Molecular Detection of blaTEM and blaCTX-M Encoding Genes from Klebsiella oxytoca Isolates from Tonsillitis. Iran J War Public Health 2023; 15 (1) :101-105
URL: http://ijwph.ir/article-1-1100-en.html
URL: http://ijwph.ir/article-1-1100-en.html
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



Rights and permissions
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
1- Department of Biology, College of Science, University of Misan, Misan, Iraq
2- Alameen Center for Advanced Research and Biotechnology, Imam Ali Holy Shrine, Najaf, Iraq
3- Department of Medical Laboratory Techniques, Faculty of Health and Medical Techniques, University of Alkafeel, Najaf, Iraq
2- Alameen Center for Advanced Research and Biotechnology, Imam Ali Holy Shrine, Najaf, Iraq
3- Department of Medical Laboratory Techniques, Faculty of Health and Medical Techniques, University of Alkafeel, Najaf, Iraq
Full-Text (HTML) (425 Views)
Introduction
Inflammation of the tonsils in the back of the throat is known as tonsillitis. The tonsils, adenoids, and lingual tonsils are all commonly inflamed during a case of pharyngitis. A painful throat, fever, swollen tonsils, difficulty swallowing, and enlarged lymph nodes in the neck are the most common symptoms. Group A beta-hemolytic Streptococcus pyogenes is the most common bacterial cause of tonsillitis, however Klebsiella species are also important human pathogens that have been linked to rising morbidity rates [1]. These bacteria are prevalent in the environment and can be found in the intestines of humans and animals, as well as water and soil. Infections and medication resistance are more likely to occur in patients who already have a damaged immune system, have been exposed to several antibiotics, or have multiple chronic conditions. In hospitalized patients, Klebsiella species frequently cause bronchopneumonia, UTIs, and septicemia [2]. They have also the ability to cause outbreaks of nosocomial infections as they often share plasmid-mediated resistance with other bacteria, which are more common at tertiary and specialized centers [3].
Among the Klebsiella spp, Klebsiella oxytoca has been isolated more frequently. K. oxytoca is a rod-shaped, nonmotile, Gram-negative bacterium with a prominent polysaccharide capsule [4-6]. Recently, K. oxytoca has emerged as one of the most antibiotic-resistant organisms responsible for outbreaks in both community and clinical settings, causing infections in patients receiving medical care. It can colonize the gastrointestinal tract, nasopharynx, and the skin, and it can cause a wide variety of infections, from relatively mild ones like a sore throat or a rash to life-threatening ones like septicemia or pneumonia [7].
Antimicrobial therapy is largely successful against infections caused by this bacteria. Antibiotics are given to people with infections to counteract the body's natural defenses. Bacteria are now resistant to -lactam antibiotics because K. oxytoca has evolved the enzymes extended spectrum -lactamases (ESBLs) and carbapenemases [8-10]. As with other Klebsiella species, K. oxytoca (formerly known as Bacterium oxytocum) may produce indole, has a positive Voges-Proskauer reaction, and liquifies gelatine. K. oxytoca is commonly picked up in the wild. Cefotaxime, ceftazidime, and aztreonam are three antibiotics that K. oxytoca is notoriously resistant to [11].
Antimicrobial sensitivity changes due to biofield treatment have been documented recently. CTX-M enzymes were reported at first time in E. coli species in 1990 [12]. These enzymes hydrolyze-actam antibiotics, leading to resistance to penicillins, cephalosporins, and aztreonam, and are encoded on plasmids, making them more horizontally transmissible [13]. Because of their greater effectiveness against cefotaxime than ceftazidime and their first isolation location (Munich; Germany), CTX-Ms are known by this acronym [14]. The CTX-M type enzymes belong to a group of class A ESBLs according Ambler classification that in general exhibit much higher levels of activity against cefotaxime and ceftriaxone than ceftazidime [15, 16].
Thus far, 172 variants of CTX-M were identified worldwide. Gram-negative bacteria are responsible for encoding the vast majority of TEM, while blaTEM-encoded genes account for about 90% of ampicillin resistance in gram-negative bacteria [17]. Single or multiple amino acid substitutions around the active site characterize the majority of TEM-type ESBLs, which are produced from mutations in the traditional TEM (TEM-1) and (TEM-2) genes via plasmid-mediated evolution. In 1965, the blaTEM-1 gene was discovered in Escherichia coli that had been isolated from a patient named Temoneira (thus, TEM) in Athens, Greece [18]. Single or several changes in the amino acid sequence of the original TEM-1 enzymes allowed for the development of TEM-2, which hydrolyzes penicillin and first-generation cephalosporins like cephaloridine [19]. These enzymes become the most commonly encountered β-lactamase among gram negative bacteria [20, 21]. Clinically, TEM-24, TEM-4, and TEM-52 are the most widely spread TEM-type ESBLs among European Enterobacteriaceae, while TEM-52, TEM-106, and TEM-116 are the most prevalent among animal isolates [22].
Because antimicrobial-resistant Klebsiella species in humans and reports on K. oxytoca are limited in the study. The aim of this study was the isolation and identification of Klebsiella oxytoca isolated from tonsillitis and detection of blaTEM and blaCTX-M in these bacteria.
Materials and Methods
Isolation and identification of bacterial isolates
After tonsillectomy, the sample surface is sterilized and opened with a sterile scalpel, and a swab is taken from the fibrosis found in the tissue to collect the sample. Fifty clinical specimens were collected from patients suffering from tonsillitis [23, 24]. These samples were collected for the period from February to September 2022 from patients from Al-Hakim Hospital, Al-Sadr Teaching Hospital, and outpatient clinics. Specimens were inoculated on routinely culture media: MacConkey agar which considered as predominant, selective and differential media for the isolation, purification and identification of K. oxytoca. As well as blood and Chocolate agar. The plates were incubated at 37˚C for 24 hours and then a single pure isolated colony was transferred to trypticase soy agar (TSA) for the preservation and to carry out other biochemical tests (IMVIC) and VITEK-2 system that confirmed the identification of isolates [25].
Specimens were inoculated on three types of culture media, including mannitol salt agar and MacConkey agar (Merk; Germany), which are considered as predominant, selective, and differential media for the isolation, purification, and identification of many types of bacteria. The plates were incubated for 24 hours at 37°C, and then a single pure isolated colony was transferred to Trypticase Soy Agar (TSA) for preservation and other biochemical tests, and the VITEK system confirmed the identification of the isolates.
DNA extraction
Genomic DNA was extracted using a commercial extraction system (Favorgen; Taiwan), according to the manufacturer's instructions.
Molecular identification
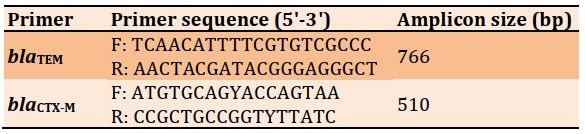
Primers used in the study were designed by Alpha DNA Company, Canada (Table 1).
Table 1) Specific primers for K. oxytoca
The PCR assay was performed to detect the blaTEM and blaCTX-M gene primers specific for K. oxytoca (Table 2). Amplified products were confirmed using 0.8% agarose gel electrophoresis to estimate the PCR product size. The gel was stained with 4µL of 0.5mg/mL ethidium bromide (Sigma; USA) and ran at 70v for 1.5h. Bands were photographed using a gel documentation system (Cleaver; UK). A 100bp ladder (Bioneer; Korea) was used to measure the molecular weights of amplified products.
Table 2) Thermal PCR program for blaTEM and blaCTX-M gene amplification in the thermocycler

Findings
Isolation and identification of K. oxytoca
The initial identification of gram negative rod was depended on the colonial morphology, biochemical tests and VITEK-2 system, 35 out of 50 of bacteria on MacConkey agar were lactose fermented produce pink colony, 15 out of 50 of isolates were lactose non fermented bacteria produce yellow or colorless colony agar, fifteen from thirty five isolates identified as Klebsiella oxytoca, other characteristic features include: production of mucoid appearance and giving indole, Methyl-red negative result but Vges-Poskeur, citrate positive result. The automated VITEK-2 compact system with GN-ID cards containing 47 biochemical tests and one negative control well was used to make the final identification. From a total of 35 possible K. oxytoca isolates, only 15 were positively identified. The confidence level of the ID message ranged from very good to excellent (probability from 95 to 99%).
Molecular detection of blaTEM and blaCTX-M encoding genes from K. oxytoca
The existence of blaTEM and blaCTX-M genes was investigated among 15 isolates of K. oxytoca, which contained 10 (66.6%) blaTEM and 11 (73.3%) blaCTX-M genes being responsible for β-lactamases. The blaTEM and blaCTX-M PCR products included 766 and 510bp, respectively (Figures 1 and 2).

Figure 1) PCR amplification products of K. oxytoca isolates amplifying the blaTEM gene product with 722bp Lane L: DNA molecular size marker (100bp ladder); Lanes isolates no. 1, 2, 3, 4, 6, 7, 9, 11, 12, 14, 15 indicate positive results for the blaTEM gene

Figure 2) PCR amplification products of K. oxytoca isolates which amplified the blaCTX-M gene product with 510 bp. Lane L: DNA molecular size marker (100bp ladder); Lanes isolates no. 1, 2, 3, 4, 6, 8, 9, 10, 11, 12, 13, 15 indicate positive results for the blaCTX-M gene
Discussion
Fifteen of 35 isolates identified Klebsiella oxytoca were lactose fermentative on MacConkey agar, produced mucoid appearance, indole positive, MR test negative result and VP, citrate test positive result, these findings agree with [30, 31]. Klebsiella growth was distinguished by its mucoid growth appearing in pink color [32, 33]. On MacConkey agar, Klebsiella colonies were lactose fermenting colonies. They gave pink color, regular edge, round, mucoid texture with large size, and K. oxytoca were (3-4mm) in diameter with a weakly mucoid aspect. The number of identified strains by the VITEK-2 in this study according to the 16-digit bionumber of laboratory reports most strain for K. oxytoca with bionumber (6707734777564010), and the other with different bionumber, although the VITEK-2 technique of automated phenotypic identification has found widespread application in clinical and scientific laboratories, it has limited performance when it comes to distinguishing between members of the K. oxytoca complex at the species level [34].
The existence of blaTEM and blaCTX-M genes was investigated among 15 isolates of K. oxytoca which contained 10 (66.6%) blaTEM and 11 (73.3%) blaCTX-M genes being responsible for β-lactamases. As shown, blaTEM and blaCTX-M shown in Figures (1) and (2), PCR products included 766 and 510 bp, respectively; this was in agreement with [28, 29]. The current study revealed that only 66.6% of the blaTEM and 73.3% of the blaCTX-M genes were found in clinical isolates, while a study by Phetburom et al. [35] exhibit blaTEM with blaCTX-M (7.72, 9.72%) and blaTEM (6.72, 8.33%). The plasmid-mediated beta lactamase TEM-1 was identified in the early 1960s. TEM-type ESBLs are a subset of this enzyme. The enzyme was named after the Greek patient Temoneira, whose blood culture contained the original strain of Escherichia coli that led to its discovery. Bacteria have evolved resistance to several of the standard antibiotics used to treat them [36].
Numerous studies have found evidence of ESBL resistance genes generated by Gram-negative bacteria. In the Asia-Pacific region, blaCTX-M and blaTEM predominated. Among the Enterobacteriaceae found in Burkina Faso, blaCTX-M (40.1%) and blaTEM (26.2%) were shown to be the most prevalent ESBL resistance genes [37]. While in Saudi Arabia the study by Ibrahim et al. [38] exhibited blaTEM (84.7%) and blaCTX-M (33.3%). The study by Alag & Aziz [39] at Maysan Province, Iraq, stated that 100% of the genes from E. coli were blaCTX-M and blaTEM. In conjunction with the present results, this shows that the prevalence of distinct ESBL gene types varies by region and even by neighborhood [40].
K. oxytoca strains isolated from humans with multidrug resistance have previously harbored four distinct blaCTX-M genes which blaCTX-M-3, blaCTX-M-9, blaCTX-M-15, and blaCTX-M-35 were isolated from different countries [41].
Conclusion
The use of the VITEK-2 system is necessary to confirm the precise identification of K. oxytoca nosocomial pathogens from tonsillitis. The existence of blaTEM and blaCTX-M gene high frequency among of K. oxytoca isolates is a concern which needs control strategies.
Acknowledgements: The authors thanks to Alameen Center for Advanced Research and Biotechnology at Imam Ali Holy Shrine, Najaf, Iraq, for their assistance in completing this study.
Ethical Permission: The studies were approved by the Human Research Ethics Committee of the University of Alkafeel, Najaf, Iraq.
Conflict of Interests: The authors declare that they have no conflict of interest.
Authors’ Contribution: Banoon ShR (First Author), Introduction Writer/Methodologist/Main Researcher/Statistical Analyst/Discussion Writer (25%); Hussein Ali Z (Second Author), Introduction Writer/Methodologist/ Main Researcher/Statistical Analyst/Discussion Writer (25%); Al-Kraety IAA (Third Author), Introduction Writer/Methodologist/Main Researcher/Statistical Analyst/Discussion Writer (25%); Aziz ZS (Fourth Author), Introduction Writer/Methodologist/Main Researcher/Statistical Analyst/Discussion Writer (25%)
Funding/Sources: There was no outside support for this study.
Inflammation of the tonsils in the back of the throat is known as tonsillitis. The tonsils, adenoids, and lingual tonsils are all commonly inflamed during a case of pharyngitis. A painful throat, fever, swollen tonsils, difficulty swallowing, and enlarged lymph nodes in the neck are the most common symptoms. Group A beta-hemolytic Streptococcus pyogenes is the most common bacterial cause of tonsillitis, however Klebsiella species are also important human pathogens that have been linked to rising morbidity rates [1]. These bacteria are prevalent in the environment and can be found in the intestines of humans and animals, as well as water and soil. Infections and medication resistance are more likely to occur in patients who already have a damaged immune system, have been exposed to several antibiotics, or have multiple chronic conditions. In hospitalized patients, Klebsiella species frequently cause bronchopneumonia, UTIs, and septicemia [2]. They have also the ability to cause outbreaks of nosocomial infections as they often share plasmid-mediated resistance with other bacteria, which are more common at tertiary and specialized centers [3].
Among the Klebsiella spp, Klebsiella oxytoca has been isolated more frequently. K. oxytoca is a rod-shaped, nonmotile, Gram-negative bacterium with a prominent polysaccharide capsule [4-6]. Recently, K. oxytoca has emerged as one of the most antibiotic-resistant organisms responsible for outbreaks in both community and clinical settings, causing infections in patients receiving medical care. It can colonize the gastrointestinal tract, nasopharynx, and the skin, and it can cause a wide variety of infections, from relatively mild ones like a sore throat or a rash to life-threatening ones like septicemia or pneumonia [7].
Antimicrobial therapy is largely successful against infections caused by this bacteria. Antibiotics are given to people with infections to counteract the body's natural defenses. Bacteria are now resistant to -lactam antibiotics because K. oxytoca has evolved the enzymes extended spectrum -lactamases (ESBLs) and carbapenemases [8-10]. As with other Klebsiella species, K. oxytoca (formerly known as Bacterium oxytocum) may produce indole, has a positive Voges-Proskauer reaction, and liquifies gelatine. K. oxytoca is commonly picked up in the wild. Cefotaxime, ceftazidime, and aztreonam are three antibiotics that K. oxytoca is notoriously resistant to [11].
Antimicrobial sensitivity changes due to biofield treatment have been documented recently. CTX-M enzymes were reported at first time in E. coli species in 1990 [12]. These enzymes hydrolyze-actam antibiotics, leading to resistance to penicillins, cephalosporins, and aztreonam, and are encoded on plasmids, making them more horizontally transmissible [13]. Because of their greater effectiveness against cefotaxime than ceftazidime and their first isolation location (Munich; Germany), CTX-Ms are known by this acronym [14]. The CTX-M type enzymes belong to a group of class A ESBLs according Ambler classification that in general exhibit much higher levels of activity against cefotaxime and ceftriaxone than ceftazidime [15, 16].
Thus far, 172 variants of CTX-M were identified worldwide. Gram-negative bacteria are responsible for encoding the vast majority of TEM, while blaTEM-encoded genes account for about 90% of ampicillin resistance in gram-negative bacteria [17]. Single or multiple amino acid substitutions around the active site characterize the majority of TEM-type ESBLs, which are produced from mutations in the traditional TEM (TEM-1) and (TEM-2) genes via plasmid-mediated evolution. In 1965, the blaTEM-1 gene was discovered in Escherichia coli that had been isolated from a patient named Temoneira (thus, TEM) in Athens, Greece [18]. Single or several changes in the amino acid sequence of the original TEM-1 enzymes allowed for the development of TEM-2, which hydrolyzes penicillin and first-generation cephalosporins like cephaloridine [19]. These enzymes become the most commonly encountered β-lactamase among gram negative bacteria [20, 21]. Clinically, TEM-24, TEM-4, and TEM-52 are the most widely spread TEM-type ESBLs among European Enterobacteriaceae, while TEM-52, TEM-106, and TEM-116 are the most prevalent among animal isolates [22].
Because antimicrobial-resistant Klebsiella species in humans and reports on K. oxytoca are limited in the study. The aim of this study was the isolation and identification of Klebsiella oxytoca isolated from tonsillitis and detection of blaTEM and blaCTX-M in these bacteria.
Materials and Methods
Isolation and identification of bacterial isolates
After tonsillectomy, the sample surface is sterilized and opened with a sterile scalpel, and a swab is taken from the fibrosis found in the tissue to collect the sample. Fifty clinical specimens were collected from patients suffering from tonsillitis [23, 24]. These samples were collected for the period from February to September 2022 from patients from Al-Hakim Hospital, Al-Sadr Teaching Hospital, and outpatient clinics. Specimens were inoculated on routinely culture media: MacConkey agar which considered as predominant, selective and differential media for the isolation, purification and identification of K. oxytoca. As well as blood and Chocolate agar. The plates were incubated at 37˚C for 24 hours and then a single pure isolated colony was transferred to trypticase soy agar (TSA) for the preservation and to carry out other biochemical tests (IMVIC) and VITEK-2 system that confirmed the identification of isolates [25].
Specimens were inoculated on three types of culture media, including mannitol salt agar and MacConkey agar (Merk; Germany), which are considered as predominant, selective, and differential media for the isolation, purification, and identification of many types of bacteria. The plates were incubated for 24 hours at 37°C, and then a single pure isolated colony was transferred to Trypticase Soy Agar (TSA) for preservation and other biochemical tests, and the VITEK system confirmed the identification of the isolates.
DNA extraction
Genomic DNA was extracted using a commercial extraction system (Favorgen; Taiwan), according to the manufacturer's instructions.
Molecular identification
Primers used in the study were designed by Alpha DNA Company, Canada (Table 1).
Table 1) Specific primers for K. oxytoca

The PCR assay was performed to detect the blaTEM and blaCTX-M gene primers specific for K. oxytoca (Table 2). Amplified products were confirmed using 0.8% agarose gel electrophoresis to estimate the PCR product size. The gel was stained with 4µL of 0.5mg/mL ethidium bromide (Sigma; USA) and ran at 70v for 1.5h. Bands were photographed using a gel documentation system (Cleaver; UK). A 100bp ladder (Bioneer; Korea) was used to measure the molecular weights of amplified products.
Table 2) Thermal PCR program for blaTEM and blaCTX-M gene amplification in the thermocycler

Findings
Isolation and identification of K. oxytoca
The initial identification of gram negative rod was depended on the colonial morphology, biochemical tests and VITEK-2 system, 35 out of 50 of bacteria on MacConkey agar were lactose fermented produce pink colony, 15 out of 50 of isolates were lactose non fermented bacteria produce yellow or colorless colony agar, fifteen from thirty five isolates identified as Klebsiella oxytoca, other characteristic features include: production of mucoid appearance and giving indole, Methyl-red negative result but Vges-Poskeur, citrate positive result. The automated VITEK-2 compact system with GN-ID cards containing 47 biochemical tests and one negative control well was used to make the final identification. From a total of 35 possible K. oxytoca isolates, only 15 were positively identified. The confidence level of the ID message ranged from very good to excellent (probability from 95 to 99%).
Molecular detection of blaTEM and blaCTX-M encoding genes from K. oxytoca
The existence of blaTEM and blaCTX-M genes was investigated among 15 isolates of K. oxytoca, which contained 10 (66.6%) blaTEM and 11 (73.3%) blaCTX-M genes being responsible for β-lactamases. The blaTEM and blaCTX-M PCR products included 766 and 510bp, respectively (Figures 1 and 2).

Figure 1) PCR amplification products of K. oxytoca isolates amplifying the blaTEM gene product with 722bp Lane L: DNA molecular size marker (100bp ladder); Lanes isolates no. 1, 2, 3, 4, 6, 7, 9, 11, 12, 14, 15 indicate positive results for the blaTEM gene

Figure 2) PCR amplification products of K. oxytoca isolates which amplified the blaCTX-M gene product with 510 bp. Lane L: DNA molecular size marker (100bp ladder); Lanes isolates no. 1, 2, 3, 4, 6, 8, 9, 10, 11, 12, 13, 15 indicate positive results for the blaCTX-M gene
Discussion
Fifteen of 35 isolates identified Klebsiella oxytoca were lactose fermentative on MacConkey agar, produced mucoid appearance, indole positive, MR test negative result and VP, citrate test positive result, these findings agree with [30, 31]. Klebsiella growth was distinguished by its mucoid growth appearing in pink color [32, 33]. On MacConkey agar, Klebsiella colonies were lactose fermenting colonies. They gave pink color, regular edge, round, mucoid texture with large size, and K. oxytoca were (3-4mm) in diameter with a weakly mucoid aspect. The number of identified strains by the VITEK-2 in this study according to the 16-digit bionumber of laboratory reports most strain for K. oxytoca with bionumber (6707734777564010), and the other with different bionumber, although the VITEK-2 technique of automated phenotypic identification has found widespread application in clinical and scientific laboratories, it has limited performance when it comes to distinguishing between members of the K. oxytoca complex at the species level [34].
The existence of blaTEM and blaCTX-M genes was investigated among 15 isolates of K. oxytoca which contained 10 (66.6%) blaTEM and 11 (73.3%) blaCTX-M genes being responsible for β-lactamases. As shown, blaTEM and blaCTX-M shown in Figures (1) and (2), PCR products included 766 and 510 bp, respectively; this was in agreement with [28, 29]. The current study revealed that only 66.6% of the blaTEM and 73.3% of the blaCTX-M genes were found in clinical isolates, while a study by Phetburom et al. [35] exhibit blaTEM with blaCTX-M (7.72, 9.72%) and blaTEM (6.72, 8.33%). The plasmid-mediated beta lactamase TEM-1 was identified in the early 1960s. TEM-type ESBLs are a subset of this enzyme. The enzyme was named after the Greek patient Temoneira, whose blood culture contained the original strain of Escherichia coli that led to its discovery. Bacteria have evolved resistance to several of the standard antibiotics used to treat them [36].
Numerous studies have found evidence of ESBL resistance genes generated by Gram-negative bacteria. In the Asia-Pacific region, blaCTX-M and blaTEM predominated. Among the Enterobacteriaceae found in Burkina Faso, blaCTX-M (40.1%) and blaTEM (26.2%) were shown to be the most prevalent ESBL resistance genes [37]. While in Saudi Arabia the study by Ibrahim et al. [38] exhibited blaTEM (84.7%) and blaCTX-M (33.3%). The study by Alag & Aziz [39] at Maysan Province, Iraq, stated that 100% of the genes from E. coli were blaCTX-M and blaTEM. In conjunction with the present results, this shows that the prevalence of distinct ESBL gene types varies by region and even by neighborhood [40].
K. oxytoca strains isolated from humans with multidrug resistance have previously harbored four distinct blaCTX-M genes which blaCTX-M-3, blaCTX-M-9, blaCTX-M-15, and blaCTX-M-35 were isolated from different countries [41].
Conclusion
The use of the VITEK-2 system is necessary to confirm the precise identification of K. oxytoca nosocomial pathogens from tonsillitis. The existence of blaTEM and blaCTX-M gene high frequency among of K. oxytoca isolates is a concern which needs control strategies.
Acknowledgements: The authors thanks to Alameen Center for Advanced Research and Biotechnology at Imam Ali Holy Shrine, Najaf, Iraq, for their assistance in completing this study.
Ethical Permission: The studies were approved by the Human Research Ethics Committee of the University of Alkafeel, Najaf, Iraq.
Conflict of Interests: The authors declare that they have no conflict of interest.
Authors’ Contribution: Banoon ShR (First Author), Introduction Writer/Methodologist/Main Researcher/Statistical Analyst/Discussion Writer (25%); Hussein Ali Z (Second Author), Introduction Writer/Methodologist/ Main Researcher/Statistical Analyst/Discussion Writer (25%); Al-Kraety IAA (Third Author), Introduction Writer/Methodologist/Main Researcher/Statistical Analyst/Discussion Writer (25%); Aziz ZS (Fourth Author), Introduction Writer/Methodologist/Main Researcher/Statistical Analyst/Discussion Writer (25%)
Funding/Sources: There was no outside support for this study.
Keywords:
References
1. Alasmari NS, Bamashmous RO, Alshuwaykan RM, Alahmari MA, Alshahrani AA, Alqarni SA, et al. Causes and treatment of tonsillitis. Egyp J Hosp Med. 2017;69(8):2975-80. [Link] [DOI:10.12816/0042838]
2. Rossolini GM, D'andrea MM, Mugnaioli C. The spread of CTX‐M‐type extended‐spectrum β‐lactamases. Clin Microbiol Infect. 2008;14 Suppl 1:33-41. [Link] [DOI:10.1111/j.1469-0691.2007.01867.x]
3. Neog N, Phukan U, Puzari M, Sharma M, Chetia P. Klebsiella oxytoca and emerging nosocomial infections. Curr Microbiol. 2021;78(4):1115-23. [Link] [DOI:10.1007/s00284-021-02402-2]
4. Doud MS, Grimes-Zeppegno R, Molina E, Miller N, Balachandar D, Schneper L, et al. A k2A-positive Klebsiella pneumoniae causes liver and brain abscess in a Saint Kitt's man. Int J Med Sci . 2009;6(6):301-4. [Link] [DOI:10.7150/ijms.6.301]
5. Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL. The function of wzy_K1 (magA), the serotype K1 polymerase gene in Klebsiella pneumoniae cps gene cluster. J Infect Dis. 2010;201(8):1268-9. [Link] [DOI:10.1086/652183]
6. Saha R, Farrance CE, Verghese B, Hong S, Donofrio RS. Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Curr Microbiol. 2013;66(1):72-8. [Link] [DOI:10.1007/s00284-012-0245-x]
7. Lin WP, Wang JT, Chang SC, Chang FY, Fung CP, Chuang YC, et al. The antimicrobial susceptibility of Klebsiella pneumoniae from community settings in Taiwan, a trend analysis. Sci Rep. 2016;6(1):36280. [Link] [DOI:10.1038/srep36280]
8. Lowe C, Willey B, O'Shaughnessy A, Lee W, Lum M, Pike K, et al. Outbreak of extended-spectrum β-lactamase-producing Klebsiella oxytoca infections associated with contaminated handwashing sinks. Emerg Infect Dis. 2012;18(8):1242-7. [Link] [DOI:10.3201/eid1808.111268]
9. Vergara-López S, Domínguez MC, Conejo MC, Pascual Á, Rodríguez-Baño J. Wastewater drainage system as an occult reservoir in a protracted clonal outbreak due to metallo-β-lactamase-producing Klebsiella oxytoca. Clin Microbiol Infect. 2013;19(11):E490-8. [Link] [DOI:10.1111/1469-0691.12288]
10. Moradigaravand D, Martin V, Peacock SJ, Parkhill J. Population structure of multidrug-resistant Klebsiella oxytoca within hospitals across the United Kingdom and Ireland identifies sharing of virulence and resistance genes with K. pneumoniae. Genome Biol Evol. 2017;9(3):574-87. [Link] [DOI:10.1093/gbe/evx019]
11. Nathisuwan S, Burgess DS, Lewis JS. Extended‐spectrum β‐lactamases: epidemiology, detection, and treatment. Pharmacotherapy. 2001;21(8):920-8. [Link] [DOI:10.1592/phco.21.11.920.34529]
12. Smet A, Van Nieuwerburgh F, Vandekerckhove TT, Martel A, Deforce D, Butaye P, e al. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. Plos One. 2010;5(6):e11202. [Link] [DOI:10.1371/journal.pone.0011202]
13. Reinthaler FF, Feierl G, Galler H, Haas D, Leitner E, Mascher F, et al. ESBL-producing E. coli in Austrian sewage sludge. Water Res. 2010;44(6):1981-5. [Link] [DOI:10.1016/j.watres.2009.11.052]
14. Birbrair A, Frenette PS. Niche heterogeneity in the bone marrow. Ann N Y Acad Sci. 2016;1370(1):82-96. [Link] [DOI:10.1111/nyas.13016]
15. Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr Issues Mol Biol. 2015;17(1):11-21. [Link]
16. Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci. 2015;22(1):90-101. [Link] [DOI:10.1016/j.sjbs.2014.08.002]
17. Livermore DM. beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8(4):557-84. [Link] [DOI:10.1128/CMR.8.4.557]
18. Steward CD, Wallace D, Hubert SK, Lawton R, Fridkin SK, Gaynes RP, et al. Ability of laboratories to detect emerging antimicrobial resistance in nosocomial pathogens: a survey of project ICARE laboratories. Diagn Microbiol Infect Dis. 2000;38(1):59-67. [Link] [DOI:10.1016/S0732-8893(00)00161-9]
19. Zhao WH, Hu ZQ. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit Rev Microbiol. 2013;39(1):79-101. [Link] [DOI:10.3109/1040841X.2012.691460]
20. Sharma J, Sharma M, Ray P. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132(3):332-6. [Link]
21. Aziz ZS, Shakir A, Addoos SA. Identification of ESBL CTX-M-15 genes from isolates of urinary tract infections. Al-Kufa Univ J Biol. 2015;7(1). [Link]
22. Coque TM, Baquero F, Cantón R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008;13(47):19044. [Link] [DOI:10.2807/ese.13.47.19044-en]
23. Shishegar M, Ashraf MJ. Posttonsillectomy bacteremia and comparison of tonsillar surface and deep culture. Adv Prev Med. 2014;2014:161878. [Link] [DOI:10.1155/2014/161878]
24. Almayali EJ, AL-Kraety IA. Molecular detection of aap gene in Staphylococcus aureus isolated from tonsillitis. Plant Arch. 2019;19:1400-2. [Link]
25. Hoffmann KM, Deutschmann A, Weitzer C, Joainig M, Zechner E, Högenauer C, et al. Antibiotic-associated hemorrhagic colitis caused by cytotoxin-producing Klebsiella oxytoca. Pediatrics. 2010;125(4):e960-3. [Link] [DOI:10.1542/peds.2009-1751]
26. Sambrook J. Molecular cloning: a laboratory manual. 3rd Edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Link]
27. Stephenson FH. Calculations for molecular biology and biotechnology. 3rd Edition. Academic Press; 2016. [Link]
28. Yang X, Lai Y, Li C, Yang J, Jia M, Sheng J. Molecular epidemiology of Pseudomonas aeruginosa isolated from lower respiratory tract of ICU patients. Braz J Biol. 2021;81(2):351-60. [Link] [DOI:10.1590/1519-6984.226309]
29. Weinroth MD, Scott HM, Norby B, Loneragan GH, Noyes NR, Rovira P, et al. Effects of ceftiofur and chlortetracycline on the resistomes of feedlot cattle. Appl Environ Microbiol. 2018;84(13):e00610-18. [Link] [DOI:10.1128/AEM.00610-18]
30. Colle JG. Mackie & Mccartney practical medical microbiology. 14th Edition. Churchill Living Stone New York: Elsevier; 1996. [Link]
31. MacFaddin JF. Biochemical tests for identification of medical bacteria. 3rd Edition. Philadelphia, PA: Lippincott Williams & Wilkins; 2000. [Link]
32. Brisse S, Grimont F, Grimont PA. The genus klebsiella. Prokaryotes. 2006:159-96. [Link] [DOI:10.1007/0-387-30746-X_8]
33. Rawy DK, El-Mokhtar MA, Hemida SK, Askora A, Yousef N. Isolation, characterization and identification of Klebsiella pneumoniae from assiut university hospital and sewage water in assiut governorate, Egypt. Assiut Univ J Botany Microbiol. 2020;49(2):60-76. [Link] [DOI:10.21608/aunj.2020.221181]
34. Yang J, Long H, Hu Y, Feng Y, McNally A, Zong Z. Klebsiella oxytoca complex: update on taxonomy, antimicrobial resistance, and virulence. Clin Microbiol Rev. 2022;35(1):e0000621. [Link] [DOI:10.1128/CMR.00006-21]
35. Phetburom N, Boueroy P, Chopjitt P, Hatrongjit R, Nuanualsuwan S, Kerdsin A. Phenotypic and molecular characterization of β-lactamase and plasmid-mediated quinolone resistance genes in Klebsiella oxytoca isolated from slaughtered pigs in Thailand. Vet World. 2022;15(2):309-15. [Link] [DOI:10.14202/vetworld.2022.309-315]
36. Humphries R, Bobenchik AM, Hindler JA, Schuetz AN. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100. J Clin Microbiol. 2021;59(12):e0021321. [Link] [DOI:10.1128/JCM.00213-21]
37. Kpoda DS, Ajayi A, Somda M, Traore O, Guessennd N, Ouattara AS, et al. Distribution of resistance genes encoding ESBLs in Enterobacteriaceae isolated from biological samples in health centers in Ouagadougou, Burkina Faso. BMC Res Notes. 2018;11(1):471. [Link] [DOI:10.1186/s13104-018-3581-5]
38. Ibrahim ME, Algak TB, Abbas M, Elamin BK. Emergence of bla TEM, bla CTX M, bla SHV and bla OXA genes in multidrug resistant Enterobacteriaceae and Acinetobacter baumannii in Saudi Arabia. Exp Ther Med. 2021;22(6):1450. [Link] [DOI:10.3892/etm.2021.10885]
39. Alag RN, Aziz ZSD. Occurrence of plasmid encoded ESBLs blaCTX-M, blaTEM genes of E. coli isolated from Clinical cases in Maysan province. Int J Sci Technol Res. 2019;8(11):122-6. [Link]
40. Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, et al. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV-and CTX-M-type β-lactamases. Antimicrob Agents Chemother. 2003;47(11):3554-60. [Link] [DOI:10.1128/AAC.47.11.3554-3560.2003]
41. Tsuka T, Ozaki H, Saito D, Murase T, Okamoto Y, Azuma K, et al. Genetic characterization of CTX-M-2-producing Klebsiella pneumoniae and Klebsiella oxytoca associated with bovine mastitis in Japan. Front Vet Sci. 2021;8:659222. [Link] [DOI:10.3389/fvets.2021.659222]










