Volume 14, Issue 2 (2022)
Iran J War Public Health 2022, 14(2): 189-195 |
Back to browse issues page
Article Type:
Subject:
History
Received: 2023/03/1 | Accepted: 2022/05/30 | Published: 2022/06/10
Received: 2023/03/1 | Accepted: 2022/05/30 | Published: 2022/06/10
How to cite this article
Abdul-Rahman M, Al-Ammar N, Kreyenberg H. HLA-DQA1 and -DQB1 Alleles as Risk Factors for Acute Lymphoblastic Leukemia. Iran J War Public Health 2022; 14 (2) :189-195
URL: http://ijwph.ir/article-1-1184-en.html
URL: http://ijwph.ir/article-1-1184-en.html
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



Rights and permissions
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
1- Microbiology Department, College of Medicine, University of Basrah, Basrah, Iraq
2- MRD-Chimarism Laboratory, Stem Cell Transplantation Centre, Goethe University Hospital, Frankfurt, Germany
2- MRD-Chimarism Laboratory, Stem Cell Transplantation Centre, Goethe University Hospital, Frankfurt, Germany
Full-Text (HTML) (758 Views)
Introduction
Leukemia is a disease of the hematopoietic system and is the most common malignancy affecting children under 15 years of age. In developed countries, Acute Lymphoblastic Leukemia (ALL) accounts for a higher percentage of childhood leukemia, while other hematological malignancies like acute myeloid leukemia occur at a lower rate in addition to characteristic chromosomal abnormalities [1].
Prenatal chromosomal abnormalities are most likely the beginning of genetic processes that occur during the manufacturing of blood cells in the fetus (hematopoietic). They operate in a two-hit illness paradigm. A very small percentage of children with pre-leukemic clones progress the disease following another genetic-related event that is expected to occur after birth (postnatal) due to exposure to mutagenic factors [2].
Human Leukocyte Antigens (HLA) are heterodimeric cell surface molecules that bind short peptides derived from non-self- and self-proteins [3]. The genes coding HLA, expressed in the Major Histocompatibility Complex (MHC), are located on chromosome 6 and are widely associated with susceptibility to many diseases and are important in immune-cell-to-cell interactions [4, 5].
Antigen presentation to T lymphocytes requires the presence of HLA molecules on the cell surface. This is why tumor cells are recognized by presenting them on HLA antigens. This recognition is mediated by tumor-specific lymphocytes called cytotoxic T-cells. The absence of HLA on the cell membrane leads to leukemia relapse and escape from T-cell immunosurveillance [6]. They encode peptides that play a role in the immunological response of the host. They are vital in tissue transplantation and linked to several infectious, autoimmune, and inflammatory illnesses [7].
According to a study by Cao et al. in 2020, specific alleles of HLA class II were associated with the susceptibility to malignant tumors including acute leukemia, and HLA-DP and -DQ loci were identified as candidate susceptibility regions for Acute Myeloid Leukemia (AML) in Han Chinese [3].
The enduring suspicion that infections and immunologic response may play a role in the etiology of childhood leukemia, particularly Acute Lymphoblastic Leukemia (ALL), is now supported, albeit still indirectly, by numerous epidemiological studies. The cumulative evidence includes, for example, descriptive observations of a peculiar peak incidence at age 2–5 years for ALL in economically developed countries, clustering of cases in situations of population mixing associated with unusual patterns of personal contacts, associations with various proxy measures for immune-modulatory exposures early in life, and genetic susceptibility conferred by variation in genes involved in the immune system [8].
The association between HLA class II alleles and leukemia may be explained in many hypotheses regarding linkage disequilibrium to the MHC-linked “leukemia gene” [9].
Some hereditary and environmental variables may cause or prevent hematologic cancers. HLA as a highly polymorphic molecule has a role in disease predisposition [10].
This study aimed to compare the most frequent -DQA1 and -DQB1 alleles in ALL patients with healthy individuals and also the relationship between these alleles and disease risk and outcome.
Materials and Methods
In this case-control study conducted during 2020-2022, 50 children with acute lymphoblastic leukemia (ALL) aged between 1-15 years and 50 sex and age-matched controls were investigated. Patients were diagnosed locally by a specialist physician based on clinical, histological, and immunophenotypic procedures in the Oncology Unit of Basrah Children's Hospital, Iraq. Subjects were selected using the convenience sampling method.
The classification of ALL types into T-ALL and B-ALL was performed based on CD markers that appeared in the flow cytometry reports of the specialist. Flow cytometry was carried out on bone marrow samples using multicolor flow cytometry and ALL markers panel. Moderate to bright CD45 expression with low SSC, cCD3+, sm. CD3+, TdT+, CD34-, CD10+, CD2+, CD5+, CD7+, CD4-, CD8+ was suggestive picture of T-ALL. Dim or negative CD45 expression with low SSC, CD19+, CD10+ (moderate and heterogeneous), HLA-DR+, cCD79a+, cIgM-, CD2-, CD33-, cCD3- and MPO- was suggestive picture of B-ALL.
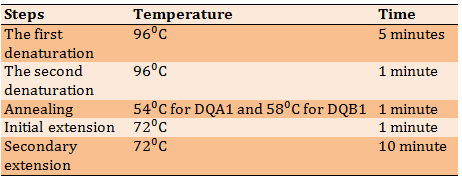
Blood samples were obtained from ALL children and controls and DNA was extracted from these samples using the Wizard Genomic DNA purification Promega Corporation kit. Electrophoresis was then performed to confirm the presence of DNA (Figure 1). The HLA-DQ gene (exon 2) was amplified using forward and reverse primers (Bioneer Corporation, USA). The primer sequence was designed according to the primers used in the previous study [11]. Sequence-based typing protocol was used to amplify the gene with some changes in annealing temperature and the final volume of Polymerase Chain Reaction (PCR). The PCR program was shown in Table 1. Gel electrophoresis and visualizing steps followed the PCR step to make sure that the amplification process was perfect. Sequencing of PCR products regarding the amplified HLA-DQA1 and HLA-DQB1 genes was sent to Korea's Macrogen sequencing laboratories. Typing of HLA alleles was done manually online by checking the sequences in the IMGT/HLA database.
Data were analyzed by SPSS 24 software using chi-square test.
Leukemia is a disease of the hematopoietic system and is the most common malignancy affecting children under 15 years of age. In developed countries, Acute Lymphoblastic Leukemia (ALL) accounts for a higher percentage of childhood leukemia, while other hematological malignancies like acute myeloid leukemia occur at a lower rate in addition to characteristic chromosomal abnormalities [1].
Prenatal chromosomal abnormalities are most likely the beginning of genetic processes that occur during the manufacturing of blood cells in the fetus (hematopoietic). They operate in a two-hit illness paradigm. A very small percentage of children with pre-leukemic clones progress the disease following another genetic-related event that is expected to occur after birth (postnatal) due to exposure to mutagenic factors [2].
Human Leukocyte Antigens (HLA) are heterodimeric cell surface molecules that bind short peptides derived from non-self- and self-proteins [3]. The genes coding HLA, expressed in the Major Histocompatibility Complex (MHC), are located on chromosome 6 and are widely associated with susceptibility to many diseases and are important in immune-cell-to-cell interactions [4, 5].
Antigen presentation to T lymphocytes requires the presence of HLA molecules on the cell surface. This is why tumor cells are recognized by presenting them on HLA antigens. This recognition is mediated by tumor-specific lymphocytes called cytotoxic T-cells. The absence of HLA on the cell membrane leads to leukemia relapse and escape from T-cell immunosurveillance [6]. They encode peptides that play a role in the immunological response of the host. They are vital in tissue transplantation and linked to several infectious, autoimmune, and inflammatory illnesses [7].
According to a study by Cao et al. in 2020, specific alleles of HLA class II were associated with the susceptibility to malignant tumors including acute leukemia, and HLA-DP and -DQ loci were identified as candidate susceptibility regions for Acute Myeloid Leukemia (AML) in Han Chinese [3].
The enduring suspicion that infections and immunologic response may play a role in the etiology of childhood leukemia, particularly Acute Lymphoblastic Leukemia (ALL), is now supported, albeit still indirectly, by numerous epidemiological studies. The cumulative evidence includes, for example, descriptive observations of a peculiar peak incidence at age 2–5 years for ALL in economically developed countries, clustering of cases in situations of population mixing associated with unusual patterns of personal contacts, associations with various proxy measures for immune-modulatory exposures early in life, and genetic susceptibility conferred by variation in genes involved in the immune system [8].
The association between HLA class II alleles and leukemia may be explained in many hypotheses regarding linkage disequilibrium to the MHC-linked “leukemia gene” [9].
Some hereditary and environmental variables may cause or prevent hematologic cancers. HLA as a highly polymorphic molecule has a role in disease predisposition [10].
This study aimed to compare the most frequent -DQA1 and -DQB1 alleles in ALL patients with healthy individuals and also the relationship between these alleles and disease risk and outcome.
Materials and Methods
In this case-control study conducted during 2020-2022, 50 children with acute lymphoblastic leukemia (ALL) aged between 1-15 years and 50 sex and age-matched controls were investigated. Patients were diagnosed locally by a specialist physician based on clinical, histological, and immunophenotypic procedures in the Oncology Unit of Basrah Children's Hospital, Iraq. Subjects were selected using the convenience sampling method.
The classification of ALL types into T-ALL and B-ALL was performed based on CD markers that appeared in the flow cytometry reports of the specialist. Flow cytometry was carried out on bone marrow samples using multicolor flow cytometry and ALL markers panel. Moderate to bright CD45 expression with low SSC, cCD3+, sm. CD3+, TdT+, CD34-, CD10+, CD2+, CD5+, CD7+, CD4-, CD8+ was suggestive picture of T-ALL. Dim or negative CD45 expression with low SSC, CD19+, CD10+ (moderate and heterogeneous), HLA-DR+, cCD79a+, cIgM-, CD2-, CD33-, cCD3- and MPO- was suggestive picture of B-ALL.
Blood samples were obtained from ALL children and controls and DNA was extracted from these samples using the Wizard Genomic DNA purification Promega Corporation kit. Electrophoresis was then performed to confirm the presence of DNA (Figure 1). The HLA-DQ gene (exon 2) was amplified using forward and reverse primers (Bioneer Corporation, USA). The primer sequence was designed according to the primers used in the previous study [11]. Sequence-based typing protocol was used to amplify the gene with some changes in annealing temperature and the final volume of Polymerase Chain Reaction (PCR). The PCR program was shown in Table 1. Gel electrophoresis and visualizing steps followed the PCR step to make sure that the amplification process was perfect. Sequencing of PCR products regarding the amplified HLA-DQA1 and HLA-DQB1 genes was sent to Korea's Macrogen sequencing laboratories. Typing of HLA alleles was done manually online by checking the sequences in the IMGT/HLA database.
Data were analyzed by SPSS 24 software using chi-square test.

Figure 1) Gel electrophoresis of genomic DNA
Table 1) The PCR program
Findings
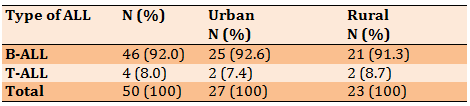
Out of 50 patients with ALL, the highest percentage was B-ALL and the lowest percentage was T-ALL. About half of subjects (54.0%) were in urban areas and 23(46.0%) were in rural areas. There was no significant relationship between the type of ALL and residency (95%CI=0.154-9.192; p>0.05; Table 2).
Table 2) Relation of B-ALL and T-ALL with residency

Findings
Out of 50 patients with ALL, the highest percentage was B-ALL and the lowest percentage was T-ALL. About half of subjects (54.0%) were in urban areas and 23(46.0%) were in rural areas. There was no significant relationship between the type of ALL and residency (95%CI=0.154-9.192; p>0.05; Table 2).
Table 2) Relation of B-ALL and T-ALL with residency

Out of 100 PCR products sent for sequencing, 60 PCR products (30 samples from patients and 30 samples from controls) were genotyped for DQA1 and DQB1.
Two alleles of DQA1 including 0201 allele and 040101 allele showed high frequency. 0201 allele was present in 13.33% of ALL patients, while it was absent in all 30 controls (p=0.05, OD=0.867) and 040101 allele was present in 23.33% of controls and present only in 3.33% of ALL patients (p=0.026, OD=0.113). One allele among HLA-DQA1 alleles appeared in IMTG/HLA during the matching of our sequences with the database as a new variant (03*new) with the frequency of 10%, but had no statistically significant relationship (p>0.05), although it was absent in all 30 controls (OD=0.90) (Table 3).
Regarding DQB1, two alleles showed significant association with ALL but in the opposite way. The allele 030201showed high frequency (33.33%) in controls (p=0.001, OD=1.500), while 060301 indicated high frequency (20%) in ALL patients (p=0.05, OD=7.250) (Table 4).
Among the results of DQB1, two alleles showed new variants and were present in both patients and controls (03*new and 06*new) (Figures 2-5).
Two alleles of DQA1 including 0201 allele and 040101 allele showed high frequency. 0201 allele was present in 13.33% of ALL patients, while it was absent in all 30 controls (p=0.05, OD=0.867) and 040101 allele was present in 23.33% of controls and present only in 3.33% of ALL patients (p=0.026, OD=0.113). One allele among HLA-DQA1 alleles appeared in IMTG/HLA during the matching of our sequences with the database as a new variant (03*new) with the frequency of 10%, but had no statistically significant relationship (p>0.05), although it was absent in all 30 controls (OD=0.90) (Table 3).
Regarding DQB1, two alleles showed significant association with ALL but in the opposite way. The allele 030201showed high frequency (33.33%) in controls (p=0.001, OD=1.500), while 060301 indicated high frequency (20%) in ALL patients (p=0.05, OD=7.250) (Table 4).
Among the results of DQB1, two alleles showed new variants and were present in both patients and controls (03*new and 06*new) (Figures 2-5).
Table 3) HLA-DQA1 alleles in ALL patients and controls
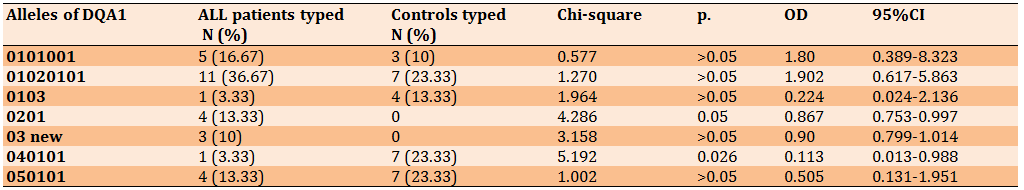
Table 4) HLA-DQB1 alleles in ALL patients and controls
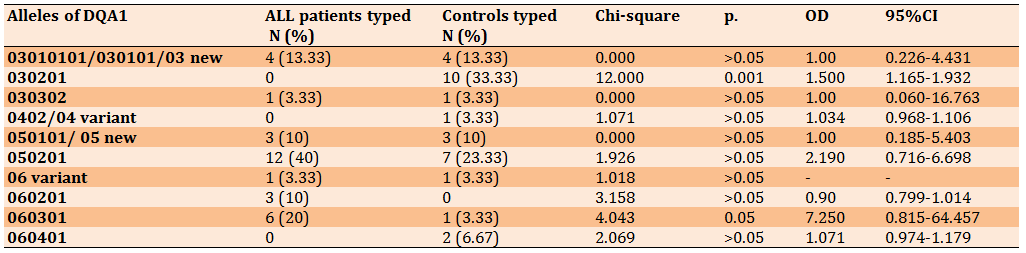

Figure 2) HLA-DQA1 base-pair substitutions in ALL patients
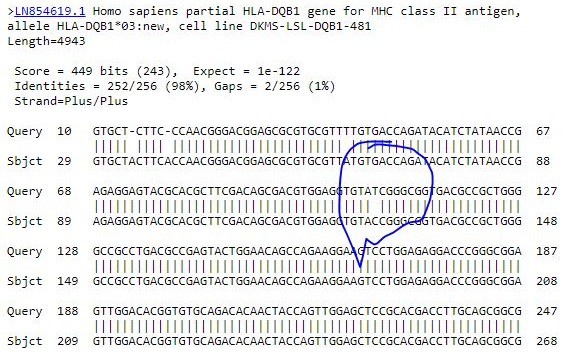
Figure 3) HLA-DQB1 base-pair substitutions in ALL patients
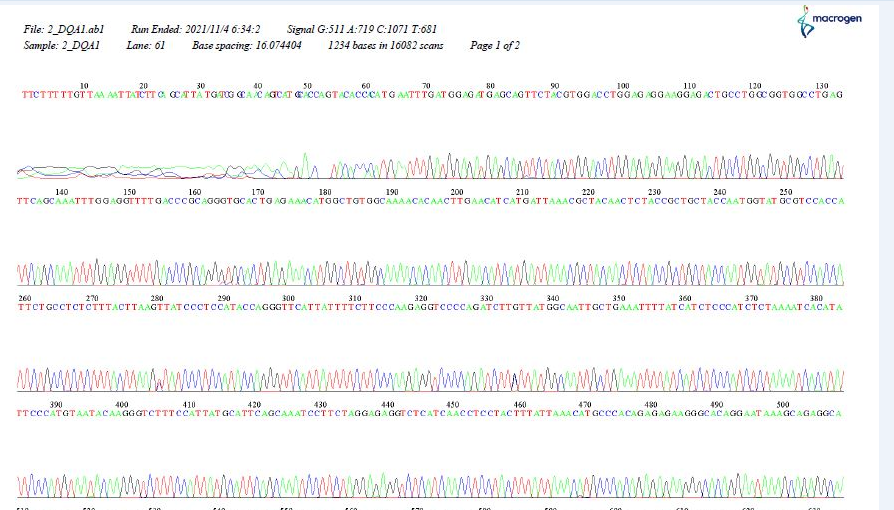
Figure 4) Chromatogram showing the sequences of HLA-DQA1
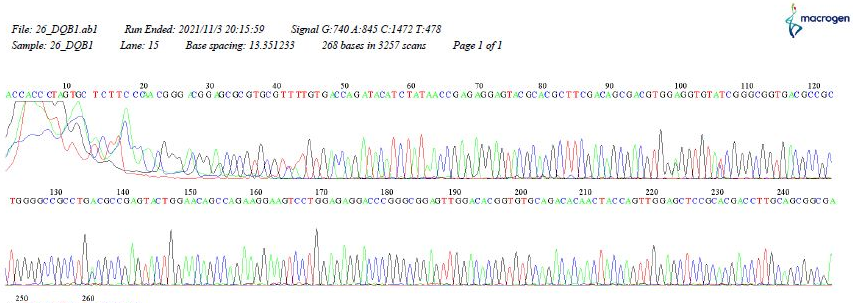
Figure 5) Chromatogram showing the sequences of HLA-DQB1
Discussion
Among oncohematological diseases, Acute Lymphoid Leukemia (ALL) and Acute Myeloid Leukemia (AML) are characterized by the uncontrolled production and accumulation of blasts that can lead to death. Although the physiopathology of these diseases is multifactorial, a genetic factor seems to be at play. Several studies worldwide have shown the association of ALL and AML with several alleles of the Major Histocompatibility Complex (MHC) [12].
Childhood leukemia is the principal subtype of pediatric cancer and, despite treatment success, its causes remain enigmatic. A plethora of candidate environmental exposures have been proposed, but most lack a biological rationale or consistent epidemiological evidence. Although there might not be a single or exclusive cause, an abnormal immune response to common infection(s) has emerged as a plausible aetiological mechanism [2].
This study aimed to compare the most frequent -DQA1 and -DQB1 alleles in ALL patients with healthy individuals and also the relationship between these alleles and disease risk and outcome.
In the present study, B-ALL showed a higher percentage than T-ALL. Residency showed no significant association with the type of leukemia.
Two alleles of DQA1 showed high frequency; 0201 allele was present in ALL patients and was absent in all controls which may indicate its role as a risk factor for ALL, while 040101 allele had a high frequency in controls and a low frequency in ALL patients.
One of the HLA-DQA1 alleles in IMTG / HLA appeared as a new variant (03*new) with 10% frequency, but had no statistically significant relationship, although it was absent in all 30 controls, further studies with a larger sample size are needed to confirm its role in ALL.
Regarding DQB1, the two alleles showed a
Among oncohematological diseases, Acute Lymphoid Leukemia (ALL) and Acute Myeloid Leukemia (AML) are characterized by the uncontrolled production and accumulation of blasts that can lead to death. Although the physiopathology of these diseases is multifactorial, a genetic factor seems to be at play. Several studies worldwide have shown the association of ALL and AML with several alleles of the Major Histocompatibility Complex (MHC) [12].
Childhood leukemia is the principal subtype of pediatric cancer and, despite treatment success, its causes remain enigmatic. A plethora of candidate environmental exposures have been proposed, but most lack a biological rationale or consistent epidemiological evidence. Although there might not be a single or exclusive cause, an abnormal immune response to common infection(s) has emerged as a plausible aetiological mechanism [2].
This study aimed to compare the most frequent -DQA1 and -DQB1 alleles in ALL patients with healthy individuals and also the relationship between these alleles and disease risk and outcome.
In the present study, B-ALL showed a higher percentage than T-ALL. Residency showed no significant association with the type of leukemia.
Two alleles of DQA1 showed high frequency; 0201 allele was present in ALL patients and was absent in all controls which may indicate its role as a risk factor for ALL, while 040101 allele had a high frequency in controls and a low frequency in ALL patients.
One of the HLA-DQA1 alleles in IMTG / HLA appeared as a new variant (03*new) with 10% frequency, but had no statistically significant relationship, although it was absent in all 30 controls, further studies with a larger sample size are needed to confirm its role in ALL.
Regarding DQB1, the two alleles showed a
significant relationship with ALL but in opposite ways. The 030201 allele showed a high frequency in the control group, which may be explained as a protective role, while the 060301 allele showed a significantly high frequency in ALL patients as a risk factor.
Among the results of DQB1, two alleles showed new variants and were present in both patients and controls (03*new and 06*new). New variants of the HLA-DQA1 and HLA-DQB1 alleles appeared during database alignment, and many base-pair substitutions were observed in some patients. This may indicate a mutation in the allele of those patients due to an error in the DNA replication process, which may require further work to confirm.
Many studies around the world have shown interest in studying the factors that may play an important role in the development of acute lymphoblastic leukemia in children. One of these factors is the major histocompatibility complex and the alleles that showed a high frequency in patients with ALL [13]. Studies on HLA alleles have shown that these alleles are considered genetic markers or can be genetically directly involved in the development of malignancies [14, 15]. Tumors can evade the immune responses by down-regulation of MHC expression; this may affect the response to treatment [16].
There are many studies on the factors associated with HLA class II gene polymorphisms [17]. An interesting study in China examined the association between HLA-DQA1 and carcinoma progression. Its results demonstrated that HLA-DQA1 plays an important role in Esophageal Squamous Cell Carcinoma (ESCC) progression and may be a biomarker for ESCC diagnosis and prognosis, as well as a potential target for the treatment of patients with ESCC [18].
A study on the Turkish population showed an increase in the frequency of specific HLA-B antigen and HLA-DRB1 alleles, this study provides evidence of a possible association between HLA molecules and acute leukemia in Turkish patients [19].
An Iranian study compared the results of HLA-DRB1 typing between patients with ALL and healthy individuals and showed an association between these alleles and ALL [20].
In our study, many new variants of HLA-DQA1 and HLA-DQB1 alleles appeared during database alignment, and many base-pair substitutions were noticed in some patients. This may indicate a mutation in those patients due to an error in the DNA replication process, which may require further work to confirm. The HLA-DQA1 allele (0201 and 03*new) and the HLA-DQB1 allele (060301) appear to be important factors in genetic susceptibility to ALL.
New variants that appeared in all patients and controls during HLA-DQB1 typing (03*new and 06*new) require more work with larger sample sizes to confirm their presence in the Basrah population and with many environmental factors facing people in Basrah to develop malignancies.
Conclusion
The HLA-DQA1 alleles (0201 and 03*new) and the HLA-DQB1 allele (060301) have a high frequency in ALL Patients. Also, new variants have appeared in all patients and controls during HLA-DQB1 typing (03*new and 06*new).
Acknowledgments: We would like to thank all the patients who participated in the current study.
Ethical Permission: 030403.
Conflict of Interests: None declared.
Authors’ Contribution: Abdul-Rahman M T (First author), Introduction Writer/Methodologist (25%); Al-Ammar N S (Second author), Introduction Writer/Methodologist/Original researcher/Statistical analyst/Discussion Writer (50%); Kreyenberg H (Third author), Introduction Writer/Statistical Analyst/ Discussion Writer (25%)
Funding: None declared.
Among the results of DQB1, two alleles showed new variants and were present in both patients and controls (03*new and 06*new). New variants of the HLA-DQA1 and HLA-DQB1 alleles appeared during database alignment, and many base-pair substitutions were observed in some patients. This may indicate a mutation in the allele of those patients due to an error in the DNA replication process, which may require further work to confirm.
Many studies around the world have shown interest in studying the factors that may play an important role in the development of acute lymphoblastic leukemia in children. One of these factors is the major histocompatibility complex and the alleles that showed a high frequency in patients with ALL [13]. Studies on HLA alleles have shown that these alleles are considered genetic markers or can be genetically directly involved in the development of malignancies [14, 15]. Tumors can evade the immune responses by down-regulation of MHC expression; this may affect the response to treatment [16].
There are many studies on the factors associated with HLA class II gene polymorphisms [17]. An interesting study in China examined the association between HLA-DQA1 and carcinoma progression. Its results demonstrated that HLA-DQA1 plays an important role in Esophageal Squamous Cell Carcinoma (ESCC) progression and may be a biomarker for ESCC diagnosis and prognosis, as well as a potential target for the treatment of patients with ESCC [18].
A study on the Turkish population showed an increase in the frequency of specific HLA-B antigen and HLA-DRB1 alleles, this study provides evidence of a possible association between HLA molecules and acute leukemia in Turkish patients [19].
An Iranian study compared the results of HLA-DRB1 typing between patients with ALL and healthy individuals and showed an association between these alleles and ALL [20].
In our study, many new variants of HLA-DQA1 and HLA-DQB1 alleles appeared during database alignment, and many base-pair substitutions were noticed in some patients. This may indicate a mutation in those patients due to an error in the DNA replication process, which may require further work to confirm. The HLA-DQA1 allele (0201 and 03*new) and the HLA-DQB1 allele (060301) appear to be important factors in genetic susceptibility to ALL.
New variants that appeared in all patients and controls during HLA-DQB1 typing (03*new and 06*new) require more work with larger sample sizes to confirm their presence in the Basrah population and with many environmental factors facing people in Basrah to develop malignancies.
Conclusion
The HLA-DQA1 alleles (0201 and 03*new) and the HLA-DQB1 allele (060301) have a high frequency in ALL Patients. Also, new variants have appeared in all patients and controls during HLA-DQB1 typing (03*new and 06*new).
Acknowledgments: We would like to thank all the patients who participated in the current study.
Ethical Permission: 030403.
Conflict of Interests: None declared.
Authors’ Contribution: Abdul-Rahman M T (First author), Introduction Writer/Methodologist (25%); Al-Ammar N S (Second author), Introduction Writer/Methodologist/Original researcher/Statistical analyst/Discussion Writer (50%); Kreyenberg H (Third author), Introduction Writer/Statistical Analyst/ Discussion Writer (25%)
Funding: None declared.
Keywords:
References
1. Wiemels J. Perspectives on the causes of childhood leukemia. Chem Biol Interact. 2012;196(3):59-67. [Link] [DOI:10.1016/j.cbi.2012.01.007]
2. Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6(3):193-203. [Link] [DOI:10.1038/nrc1816]
3. Cao S, Wu Y, Qian X, Ma H. Genetic variants in HLA-DP/DQ contribute to risk of acute myeloid leukemia: A case-control study in Chinese. Pathology Res Pract. 2020;216(3):152829. [Link] [DOI:10.1016/j.prp.2020.152829]
4. Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5(12):889-99. [Link] [DOI:10.1038/nrg1489]
5. Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38(1):75-81. [Link] [DOI:10.1038/ng1697]
6. Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181-273. [Link] [DOI:10.1016/S0065-2776(08)60911-6]
7. Gregersen JW, Kranc KR, Ke X, Svendsen P, Madsen LS, et al. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature. 2006;443:574-7. [Link] [DOI:10.1038/nature05133]
8. Urayama KY, Thompson PD, Taylor M, Trachtenberg EA, Chokkalingam AP. Genetic variation in the extended major histocombatibility complex and susceptibility to childhood acute lymphoblastic leukemia: a review of the evidence. Front Oncol. 2013;3:300. [Link] [DOI:10.3389/fonc.2013.00300]
9. Taylor M, Hussain A, Urayama K, Chokkalingam A, Thompson P, et al. The human major histocompatibility complex and childhood leukemia: an etiological hypothesis based on molecular mimicry. Blood Cells Molecules Dis. 2009;42(2):129-35. [Link] [DOI:10.1016/j.bcmd.2008.10.009]
10. Kazemi MH, Momeni-Varpashti Z, Roshandel E, Sankanian G, Hosseini N, et al. Association of HLA alleles with hematologic malignancies. Gene Rep. 2021;25:101346. [Link] [DOI:10.1016/j.genrep.2021.101346]
11. Al-Ammar NS. HLA-DQA1 and HLA-DQB1 genotyping among lichen planus patients in Basrah province. Medi J Basrah Univ. 2014;32(1):43-53. [Link] [DOI:10.33762/mjbu.2014.94512]
12. Fernandez-Torres J, Flores-Jimenez D, Arroyo-Perez A, Granados J, Lopez-Reyes A. HLA-B*40 allele plays a role in development of acute leukemia in Mexican population: a case-control study. Biomed Res Int. 2013;2013:705862. [Link] [DOI:10.1155/2013/705862]
13. Graubert TA, Mardis ER. Genomics of acute myeloid leukemia. Cancer J. 2011;17(6):478-91. [Link] [DOI:10.1097/PPO.0b013e31823c5652]
14. Haouas H, Haouas S, Uzan G, Hafsia A. Identification of new markers discriminating between myeloid and lymphoid acute leukemia. Hematology. 2010;15(4):193-203. [Link] [DOI:10.1179/102453310X12647083620769]
15. Bene MC, Porwit A. Acute leukemias of ambiguous lineage. Seminars Diagnos Pathol. 2012;29(1):12-8. [Link] [DOI:10.1053/j.semdp.2011.08.004]
16. Haworth KB, Leddon JL, Chen CY, Horwits EM, Mackall CL, Cripe TP. Going back to Class I: MHC and Immunotherapies for childhood cancer. Pediatr Blood Cancer. 2015;62(4):571-6. [Link] [DOI:10.1002/pbc.25359]
17. Wu J, Song Y, Chen F, Xiao H. Study on the association of the polymorphism of HLA-II gene with leukemia. Oncol Lett. 2017;14(1):224-8. [Link] [DOI:10.3892/ol.2017.6136]
18. Shen FF, Pan Y, Li JZ, Zhao F, Yang HJ, et al. High expression of HLA-DQA1 predicts poor outcome in patients with esophageal squamous cell carcinoma in Northern China. Medicine (Baltmore). 2019;98(8):e14454. [Link] [DOI:10.1097/MD.0000000000014454]
19. Ozdilli K, Oguz FS, Anak S, Kekik C, Carin M, Gedikoglu G. The frequency of HLA class I and II alleles in Turkish childhood acute leukemia patients. J Int Med Res. 2010;38(5):1835-44. [Link] [DOI:10.1177/147323001003800531]
20. Yari F, Sobhani M, Sabaghi F, Zaman-Vaziri M, Bagheri N, Talebian A. Frequencies of HLA-DRB1 in Iranian normal population and in patients with acute lymphoblastic leukemia. Arch Med Res. 2008;39(2):205-8. [Link] [DOI:10.1016/j.arcmed.2007.09.009]








